Human Microbiomes
Dr. Metcalf’s lab was recently featured in a segment narrated by Ed Yong, the acclaimed science writer who authored “I Contain Multitudes”, which lets us peek into the ubiquitous nature of microbes and how vital they are.
A microbial clock to estimate the time since death
Researcher: Jessica L. Metcalf
Estimating the time since death is critical in death investigations because it can help with reconstructing crime scenes, issuing of death certificates, and validating alibis. The time since death can be estimated using a range of methods including last communications as well as biological clues such as rigor mortis (stiffening of muscles) or forensic entomology (study of insects). However, each approach has limitations and new methods are needed. Microbes have excellent potential to be used as physical evidence because they are ubiquitous and have predictable ecologies. In collaboration UCSD, Chaminade University and several other institutions, I have been developing a new method that uses the succession of microbial decomposer communities after death as an indicator of time since death. Using DNA sequencing of cadaver-associated skin or soil microbes, we can predict the time since death with an accuracy of +/-3 days over 21 days, which is a significant advance for the field.
caption text
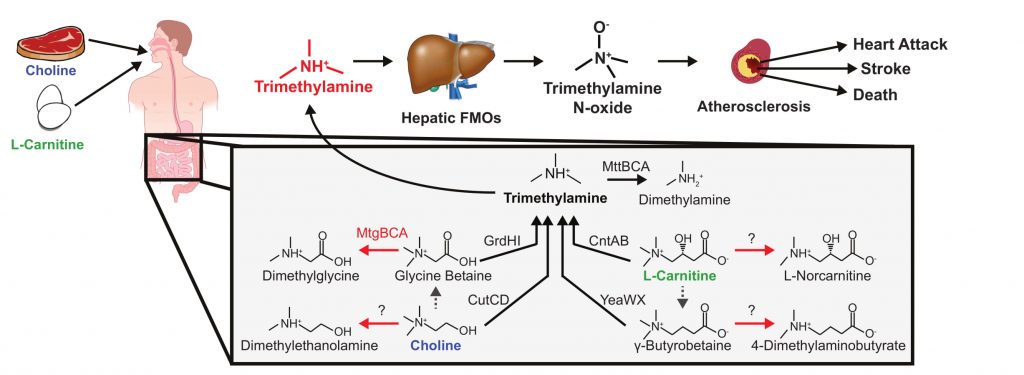
Uncovering novel pathways for methylated amine utilization in the gut
Researcher: Kelly C. Wrighton
The Wrighton lab is investigating the link between the gut microbiome and cardiovascular disease in humans. Cardiovascular disease is the leading cause of mortality in the United States and evidence is accumulating that certain human intestinal microbes contribute to atherosclerosis leading to cardiovascular disease. Recent reports have shown that gut microbes convert quaternary amines like choline and carnitine into trimethylamine. This trimethylamine then enters the bloodstream and is converted by liver flavoproteins to trimethylamine-N-oxide (TMAO), a trigger for atherosclerosis. Trimethylamine production has long been considered the sole route of microbial quaternary amine degradation under the anaerobic conditions prevalent in the gut; but recent evidence reveals a more complex microbial network. Intestinal isolates have been shown to demethylate quaternary amines during growth- these reactions would not generate trimethylamine, thereby reducing the risk for cardiovascular disease. The central hypothesis of this work is that quaternary amine demethylation might serve to moderate microbial trimethylamine production in the human intestine, providing a mechanism for homeostasis or therapeutic control of TMAO levels. To investigate the prevalence and activity of these pathways in the human gut, fecal and urine samples along with subject data were collected from a cohort of 150 people. The metabolome of urine and fecal samples coupled to metagenomes and metatranscriptomes from fecal samples were obtained. Ongoing investigation is piecing together the pathways for methylated amine utilization in the human gut.
caption text
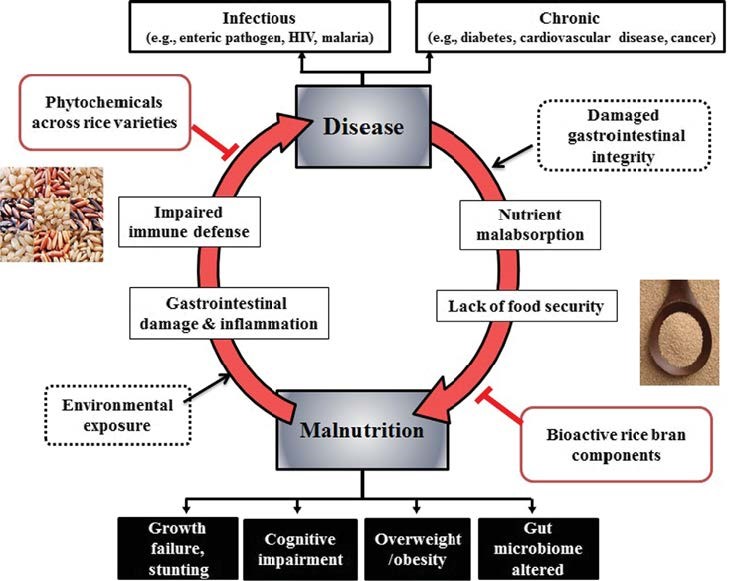
Dietary interactions with human microbiomes across the lifespan
Researcher: Elizabeth Ryan
Diarrhea and malnutrition represent leading causes of death in developing countries, and practical, low cost interventions are critical to ensure the success of prevention and treatment. Based on findings of heat stabilized rice bran to 1) supply plant lipids, vitamins and amino acids, 2) enhance gut mucosal immunity, 3) promote growth and metabolism by native gut probiotics and 4) reduce pathogen induced diarrhea in animals, our team conducted clinical trials in infants during weaning. This project is focused on evaluating changes in the developing infant microbiome composition and function in response to rice bran consumption. We identified a complete country-level separation in the overall gut microbial community composition, and have proceeded to identify rice bran responsive taxa for that are associated with growth outcomes at 8 and 12 months of age. Lactobacillus, Lachnospiraceae, Bifidobacterium, Ruminococcaceae and Veillonella were examples of taxa identified as responsive following rice bran consumption compared to an age and control matched group. We are interested to evaluate microbe-metabolite interactions related to saccharolytic mechanisms of action, crossfeeding as well as competitive inhibition of pathogen colonization and antimicrobial resistance. The stool metabolome was also analyzed alongside microbial communities for assessing how gut microbes metabolize rice bran components. These studies were supported by Grand Challenges Exploration grant in Global Health from the Bill and Melinda Gates Foundation.
Microbial metabolism of Rice Bran for colon cancer control and prevention
Colorectal cancer (CRC) develops as a result of complex interactions between the host mucosal immune response, microbiota, diet components, colon cancer stem cells (CSC) and inflammatory mediators. Rice bran is a `wasted’ agricultural byproduct of rice processing that has shown CRC control and prevention efficacy in numerous animal studies. Our metabolomics investigations revealed novel probiotic metabolism of rice bran, and our completed dietary feeding studies in adult colon cancer survivors provide compelling support that rice bran is a promising, novel dietary agent to be metabolized by the gut microbiota in favor of reducing CRC risk. Rice bran is a unique source of plant lipids and contains distinct contents from wheat, oat and other cereal grains. The current project investigates rice bran phytochemicals and the microbial digested byproducts of rice bran for enhancing gut mucosal immunity, decreasing colonic inflammation, and reducing neoplastic growth. Microbes from humans that consumed rice bran are being tested in germ free animal models of colon cancer progression to identify whether the microbes and/or the metabolite products are effective. The major objective of this proposal is to fill a significant gap in our knowledge regarding the microbiome-mediated mechanisms of action for rice bran in proximal and distal colon cancer prevention. This work is funded by NIH (1R01CA201112-01A1).