Environmental Microbiome
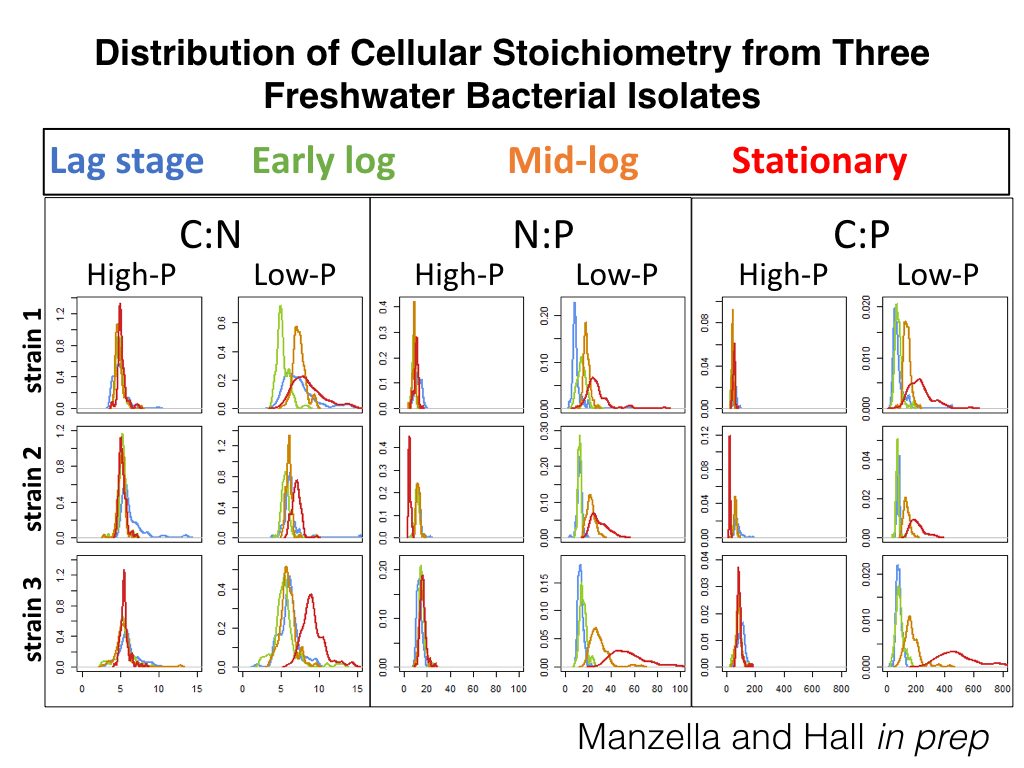
Evaluating the Stoichiometric Trait Distribution of the Microbiome
The Hall lab is interested in how different cues affect the biomass composition of bacterial populations and communities. One way to evaluate biomass composition is to evaluate the ratios of elements (i.e., stoichiometry) that compose a microbe’s biomass. The ecological stoichiometry of microbial biomass has most often focused on the ratio of the biologically-important elements carbon (C), nitrogen (N), and phosphorus (P) and has primarily been examined at a resolution where the contribution of the individual is masked by the reported population or community average. However, reporting population or community averages makes it difficult to assess phenotypic plasticity and may mask important information to understand both the drivers and implications of microbial biomass stoichiometry in nature. One way to assess the diversity of individual microbial phenotypes is through the use of single-cell techniques such as energy dispersive spectroscopy (EDS). EDS reports cellular quotas for the majority of elements composing microbial biomass including C, N, and P. By measuring individual cells within a microbial community or population, the stoichiometry of microbial biomass can then be described as a distribution instead of an average. To assess stoichiometric variance within populations and communities we measure the stoichiometric trait distribution of freshwater bacterial populations under different resource and environmental stressor conditions and compared them to natural microbial communities sampled from lakes or streams. Exploration of stoichiometric trait distributions has the potential to improve our understanding of how nutrients interact with organisms to structure the elemental content of bacterial biomass and better describe how microbiome biomass composition affects ecosystems.
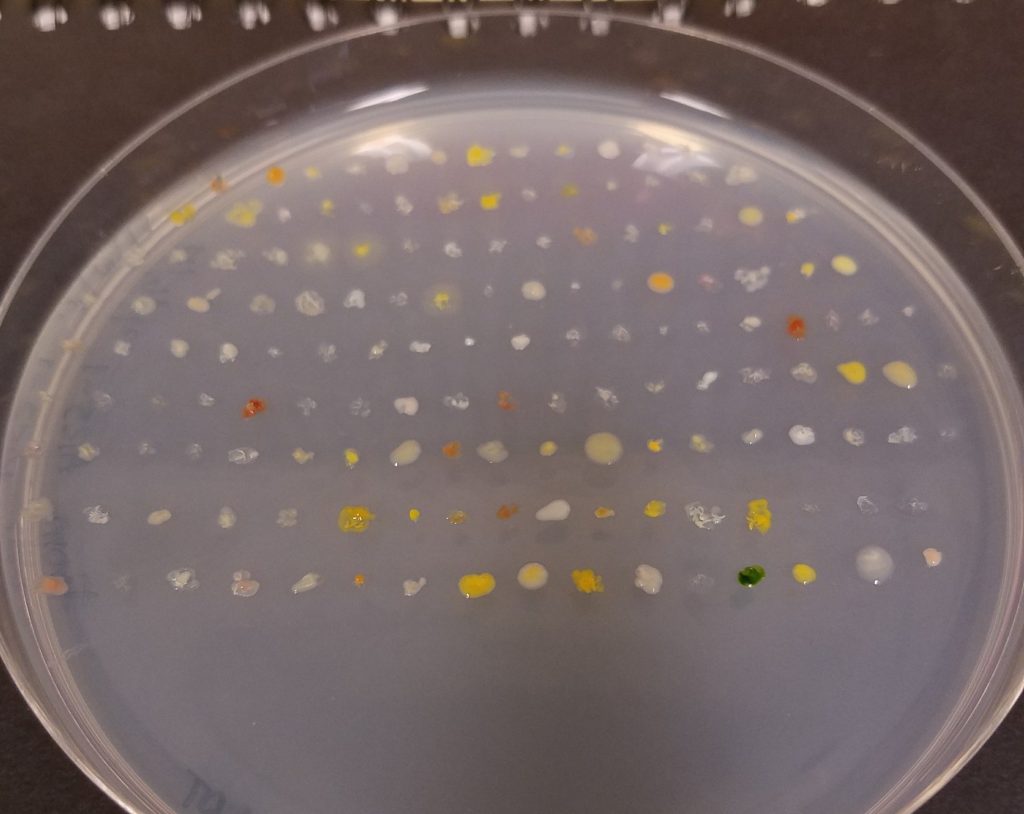
How Stream Microbiomes Influence Stream Food Webs
The Hall lab studies the microbiomes of stream ecosystems. The upper Arkansas River has been impaired by metal pollution due to historical mining in nearby Leadville, Colorado. By the late 1990s, implementation of water treatment facilities and various habitat enhancements resulted in significant improvements in water quality. Despite improved water quality, differences remain in benthic macroinvertebrate community structure between upstream and downstream sites – greater relative abundance of metal-sensitive taxa (e.g. mayflies) at reference upstream sites and more metal-tolerant taxa (e.g. caddisflies) at sites downstream of California Gulch, the inlet that contributes heavy metal contamination to the upper Arkansas River. In collaboration with Will Clements, from the Department of Fish and Wildlife, and Brian Wolff, a PhD candidate in the Graduate Degree Program in Ecology at CSU, we hypothesized that the observed difference in macroinvertebrate community structure may be due to how metals alter biomass composition of the stream microbiome. This change in the base of the stream food web would indirectly structure macroinvertebrate communities through altered food quality (i.e., microbial biomass). We sampled benthic sediments at multiple sites upstream and downstream of California Gulch, across multiple seasons (spring and fall) and years (2015 – 2017). Our results showed: microbial communities were indeed different between sites upstream and downstream of California Gulch, metals were primarily responsible for any observed difference in community structure, and there were differences in the metal content and biomass stoichiometry of the stream microbiome between sites in the river. We also isolated >300 bacteria from sediments at each site on media containing Copper and Zinc. We are in the process of comparing how the biomass composition is altered when cells are grown in media containing metals compared to the biomass composition of cells grown in the absence of metals. This is one example of how combining mechanistic physiological research and ecosystem scale observational research we hope to elucidate how microbiomes influences the ecosystems they inhabit.